MIPT determines a molecular mechanism of anti-cancer therapeutic candidate
An international team of scientists, including MIPT researchers, has defined the way their promising anti-cancer molecules work. The research findings will help to further optimize these new agents in order to develop drug candidates that are effective and safe for healthy tissue. The study was published in Bioorganic & Medicinal Chemistry.
Many of the current anti-cancer treatments destroy healthy cells along with fighting a tumor. To reduce or eliminate these undesirable effects, we need to better understand how drugs work and what their molecular targets are.
The scientists studied the influence of a number of compounds called thienopyridines on sea urchin embryos and a panel of human cancer cells. In parallel, they used molecular modeling to perform a detailed analysis of the interaction between the anti-tumor agent and specific targets in cells. It was established in prior studies that thienopyridines are able to inhibit cancer cell growth; however, the precise biological mechanisms by which they affect cells remained unknown.
"Our study unequivocally demonstrated that our new small molecules bind microtubules. Moreover, by using molecular modeling, we were able to pinpoint the spot on the tubulin molecule where this binding occurs. The resulting data can be used to make the anti-cancer molecule more potent, selective and suitable for testing in tumor models," comments Prof. Alex Kiselyov of MIPT.
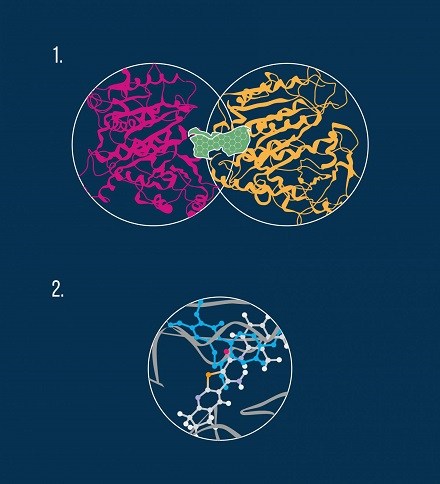
For many of the most efficacious chemotherapeutic agents, microtubule destabilization is the main mechanism of action. Microtubules are structures within a cell that play a key role in mitosis, a crucial stage in the cell division process. Chemically, a microtubule is a gigantic biological aggregate formed by protein subunits called tubulin. An anti-cancer drug can bind to at least three distinct areas, or pockets, on the microtubule, namely the colchicine site, the vinca alkaloid site, and the taxol site (see diagram).
By performing in vivo experiments, the researchers confirmed that the compounds examined in the study indeed bind to tubulin molecules and thus exhibit a destabilizing effect on microtubules. In particular, molecular modeling revealed that the anti-cancer molecules interact with the colchicine pocket (see diagram).
The algorithm used by the scientists involved several steps including identifying potential interacting sites on the tubulin dimer, prioritizing the most energetically favorable binding poses for the new agents, matching their topology to the three tubulin inhibition sites, and finally selecting the compounds that exhibit the best binding energy. In agreement with these computational findings, phenotypic in vivo data confirmed the colchicine binding site on the tubulin molecule to be the most likely target for the new microtubule-destabilizing molecules.
The scientists have been actively searching for novel anti-cancer molecules with improved activity and safety. In their previous studies, they proposed a method of synthesizing anti-tumor agents based on compounds extracted from parsley and dill seeds and found a molecule to fight chemoresistant ovarian cancer.
The team hopes that the data obtained from this research will help to optimize a series of molecules (thienopyridines) for further studies in animals to ultimately develop new anti-cancer drugs.