The recent research from the Center for Algorithmic and Robotized Synthesis at the Institute for Basic Science (IBS) in South Korea has generated skepticism within the scientific community. The study suggests that AI-predicted structures, using DeepMind’s AlphaFold2 algorithm, can indicate protein mutant stability. Despite the potentially beneficial implications of the research, many important questions have been raised about the validity and applicability of the findings.
One primary concern is whether AlphaFold2 has genuinely understood the physics of protein folding, or if it functions more as a sophisticated regression model that identifies statistical patterns. The study’s authors, John McBride, and Tsvi Tlusty, sought to assess AlphaFold2's capability to predict the effects of mutations on stability, a challenging task given the multitude of potential mutations and the intricate nature of protein folding.
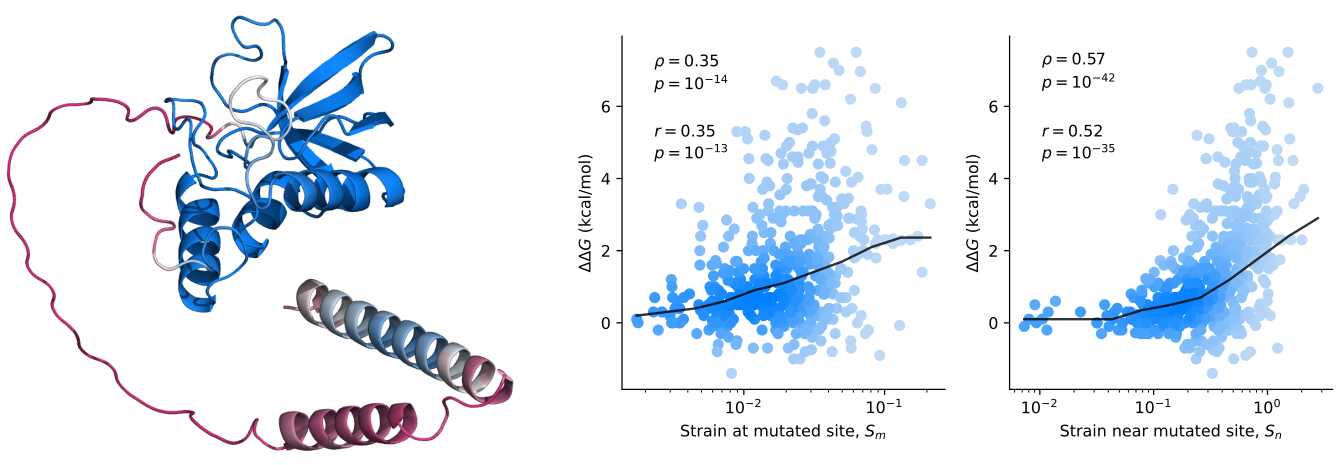
The research results propose that the structural changes anticipated by AlphaFold2 are connected to changes in stability induced by mutations. While this discovery could imply that the algorithm contains valuable stability information, critics have raised doubts about the depth and accuracy of these predictions. Lead author John McBride's assertion that the predicted structures encode significant physical information has been met with skepticism, as some experts question how AI algorithms can accurately capture the complexities of protein behavior.
Moreover, the study's use of an innovative metric called effective strain to identify subtle structural changes and their impact on stability has faced scrutiny for its methodology and potential biases. The researchers' statement that significant structural changes predicted by AlphaFold2 correspond to substantial changes in stability has been met with cautious skepticism, as protein engineering necessitates rigorous validation and reproducibility.
While the research represents a significant advance in exploring the intersection of AI and protein stability, the scientific community remains cautious about the implications for protein engineering and drug development. Scientists emphasize the critical need for thorough validation, replication, and independent verification of the findings, highlighting the importance of rigor and reliability in scientific research.
As the debate regarding the IBS research on protein mutant stability continues, the scientific community remains divided on whether AI-predicted structures can genuinely enhance our understanding of protein behavior and stability. The conclusions of the study, though intriguing, have sparked skepticism and call for a comprehensive examination of the methodology, assumptions, and implications of the research within the broader context of protein engineering and biological science.


 How to resolve AdBlock issue?
How to resolve AdBlock issue?