Researchers can now use the Browser's features to see genetic code at any scale and add annotations for global collaboration
Research into the novel Wuhan seafood market pneumonia virus, the deadly "coronavirus" that has forced the Chinese government to quarantine more than 50 million people in the country's dense industrial heartland, will be facilitated by the UC Santa Cruz Genomics Institute. The Genomics Institute's Genome Browser team has posted the complete biomolecular code of the virus for researchers all over the world to use.
"When we display coronavirus data in the UCSC Genome Browser, it lets researchers look at the virus' structure and more importantly work with it so they can research how they want to attack it," said UCSC Genome Browser Engineer Hiram Clawson.
Samples of the virus have been processed in labs all over the world, and the raw information about its genetic code has been sent to the worldwide repository of genomic information at the National Institutes of Health's National Center for Bioinformatics (NCBI) in Bethesda, Maryland. 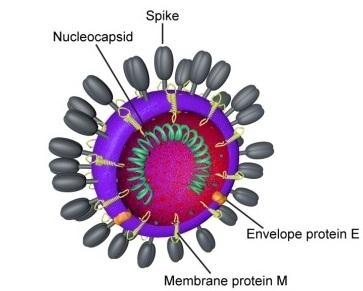 {module INSIDE STORY}
{module INSIDE STORY}
"The NCBI is a worldwide repository established in the very early days of genomics," said Clawson. "When people find novel viruses, they send them to the NCBI, and the NCBI assigns them a name and number so everyone can refer to an exact specimen. Once they've processed the genomic information, it's made available to the world from the database."
From there, the UC Santa Cruz Genome Browser processes the information into a visual display of the virus.
The NCBI named the Wuhan seafood market pneumonia virus 2019-nCoV, which stands for novel coronavirus discovered in 2019.
UC Santa Cruz retrieved the information, consisting of 29,903 nucleotides -- the base pairs that make up the DNA and RNA molecules that encode all life on earth.
"When we obtain this data from NCBI, it's a single file with the letters in it from the DNA or RNA (A, C, G, and T)," Clawson said. "This one happens to be single-stranded RNA, a relatively simple structure.
This information is processed and placed into a database, where the Genome Browser can access the material and display it in a web browser in a much more useful format.
"What makes the Genome Browser so valuable is that it is so visual," Clawson said. "It makes it very clear where everything is, so when people make interesting measurements about the genome in the virus, they can see what they're looking at," Clawson said.
Researchers can zoom in and out of the genome. This allows them to see base pairs at the most detailed level. Or, they zoom all the way out and see the 10 individual genes that the 29,903 base pairs comprise.
The Browser also contains a CRISPR track, which allows researchers to see where they can splice genetic material and how they can cut it. With CRISPR, researchers can edit the genetic material, a tremendously valuable tool for determining which genes do what.
"In the case of this virus," Clawson said, "there are approximately ten genes and the largest is its spike protein," referring to the chemical spine which the virus uses to snag onto human cells and hijack their cellular machinery to reproduce themselves. "So they might make a change to see if it makes the spike protein more or less virulent."


 How to resolve AdBlock issue?
How to resolve AdBlock issue?