The structure of a biomolecule can reveal much about its functioning and interaction with the surrounding environment. The double-helical structure of DNA and its implications for the processes of transmission of genetic information form an obvious example. In a new study by SISSA - Scuola Internazionale Superiore di Studi Avanzati, experimental data were combined with supercomputer simulations of molecular dynamics to examine the conformation of an RNA fragment involved in protein synthesis and its dependence on the salts present in the solution. The research has led to a new method for a high-resolution definition of the structures of biomolecules in their physiological environments. 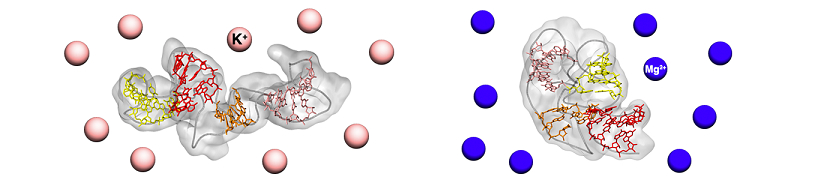
"X-ray crystallography, as used to discover the double-helical conformation of DNA, remains one of the most common techniques for studying biomolecule structures", explains SISSA physicist Giovanni Bussi. "This technique allows us to reconstruct the image of the molecule in solid-state crystalline form. However, this yields a static view of the structure that may not correspond to that assumed in the aqueous natural environment in which biomolecules are normally found."
This is why researchers began to use the small-angle X-ray scattering (SAXS) technique in the last decade to study RNA molecules, which can have highly dynamic structures. This method can be used directly in aqueous solutions that reproduce the physiological environment. Furthermore, the composition of the solutions can be modified to study how the molecules adapt to different conditions. Unfortunately, however, SAXS has limited resolution, in the order of a nanometer. Giovanni Bussi and Mattia Bernetti, a research fellow at SISSA, therefore decided to enhance SAXS via a 'computational microscope', combining it with molecular dynamics simulations that allow computerized reconstruction of molecular structures at the atomic level.
"We studied a fragment of ribosomal RNA involved in protein synthesis," explains the researchers. "We used SAXS data, derived from aqueous solutions containing different mixtures of salts, that was provided by Kathleen B. Hall of the Washington University School of Medicine in St Louis and combined them with molecular dynamics simulations. By this means we discovered the existence of two distinct conformations: one more compact and functional to the protein synthesis process, the other more extended, confirming the dynamic nature of RNA. In particular, we noticed how the prevalence of one structure over the other varies with the salts dissolved in solution, further underlining the importance of studying these molecules in an environment as similar as possible to that of the cell."
Bernetti and Bussi conclude that the results of the study, published in Nucleic Acids Research, have significance beyond the specific case and indicate an innovative method offering two advantages: "In this work, we combined molecular dynamics simulations and SAXS experimental data to obtain high-resolution structures of RNA biomolecules. This is a useful approach in two senses: on one hand, it allows detail to be added to SAXS experimental data, which in fact give a very approximate view; on the other hand, it allows results of molecular dynamics to be corrected if the models used in the simulations are insufficiently accurate."


 How to resolve AdBlock issue?
How to resolve AdBlock issue?