DEVELOPER TOOLS
Waseda University researchers develop RaptGen computational model that can be used for novel aptamer generation
Oligonucleotides are short, single strands of synthetic DNA or RNA. Albeit small, these molecules play an important role in molecular and synthetic biology applications. One type of oligonucleotide—aptamers—can selectively bind to specific targets such as proteins, peptides, carbohydrates, viruses, toxins, metal ions, and even live cells. As they are similar to antibodies, they have a variety of uses in the fields of biosensors, therapeutics, and diagnostics. However, compared to antibodies, aptamers do not induce an immune reaction in our bodies and are easy to synthesize and modify. Moreover, an aptamer’s three-dimensional folding structure allows it to bind to a wider range of targets. 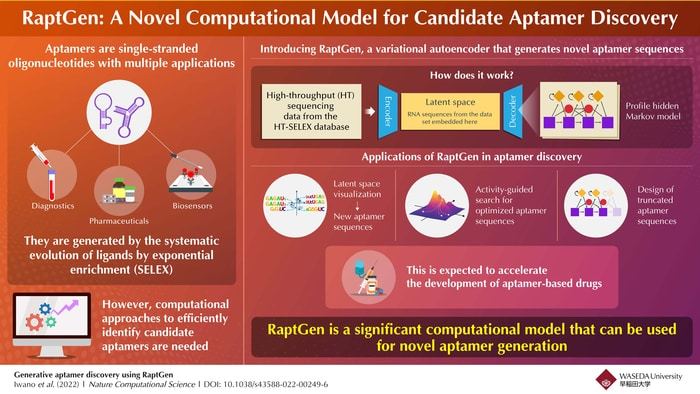
Aptamers are usually generated by an in vitro selection and amplification technology called systematic evolution of ligands by exponential enrichment, or SELEX. Briefly, SELEX is based on repeated cycles of binding, separation, and amplification of nucleotides. This process results in an enriched pool of nucleotide sequences that are then analyzed for candidate selection. High-throughput SELEX (HT-SELEX) can generate a vast number of aptamer candidates, but current practically-applicable sequencing only allows us to evaluate a limited number of these candidates (approximately 106). Therefore, computational processes are essential to optimize the discovery of new aptamers.
Variational autoencoder (VAE, a type of machine learning approach)-based compound designs have been reported to be beneficial in the discovery of other small molecules. Now, a team of researchers led by Professor Michiaki Hamada of the Graduate School of Advanced Science and Engineering in Waseda University, Japan, introduced RaptGen, a VAE that can be used for aptamer generation. In their paper, they describe how RaptGen uses a VAE with a profile hidden Markov Model decoder to create latent spaces in which sequences can form clusters. By using this latent representation, RaptGen was able to generate aptamers that were not included even in the original sequencing data or HT-SELEX dataset.
When asked how exactly RaptGen could boost aptamer discovery, Professor Hamada states, “RaptGen first visualizes a latent space with a sequence motif, then generates multiple new aptamer sequences via this latent space. For example, it searches for optimized aptamer sequences in the latent space by considering additional information after analyzing the activity of a subset of sequences. Additionally, RaptGen enables the design of shortened (or truncated) aptamer sequences.”
The team also successfully evaluated RaptGen’s performance using real-world data, by subjecting it to data from two independent HT-SELEX datasets. RaptGen could generate aptamer derivatives in an activity-guided manner and provide opportunities to optimize their activities. “This is important as it means that RaptGen can generate sequences having desired properties, such as the inhibition of certain enzymes or protein-protein interactions,” Professor Hamada explains. The application of these molecules could open many doors in the future.
Moving forward, the team plans to conduct extensive studies evaluating if alternative models can improve the performance of RaptGen and whether RaptGen could advance RNA aptamer generation by using RNA sequences. The only drawbacks of using RaptGen are the high computational cost and increased training time, both of which can be improved in further studies.
Professor Hamada summarizes by saying, “To the best of our knowledge, RaptGen is the only data-driven method that can design and optimize truncated aptamers directly from HT-SELEX data. We believe that in due time, RaptGen will be recognized as a key tool for efficient aptamer discovery.”
Here’s to their vision of a healthy and long-lived society with better therapeutics!
