Laboratory research is the possibly best-known driving force for advancement in science. However, when it comes to investigating evolutionary processes, lab work often faces its limitations. This is where the power of big data and computational modeling comes into play. A recent joint effort from the Bielefeld University and the Leibniz Institute of Plant Genetics and Crop Plant Research (IPK) in Gatersleben to understand the evolution of C4 photosynthesis, demonstrates the potential of in silico modeling in science. Using constraint-based modeling, the researchers were able to step back in time and predict the evolutionary pathway of this particular form of photosynthesis.
All plants, algae and select bacteria perform photosynthesis, converting water and carbon dioxide (CO2) into glucose with the help of energy from sunlight. As they all produce their own food, they are classed as autotroph organisms. However, the process of photosynthesis is not the same in all autotrophs. The most common type of photosynthesis in plants is the C3 photosynthesis. It relies on the enzyme Rubisco for the fixation of CO2. Despite its prevalence in the plant-world, the C3 process has its downsides, as the function of Rubisco is slow and also unspecific. Instead of fixing CO2, plants can accidentally fix oxygen, thus producing toxic by-products that need to be recycled. To avoid these detrimental aspects, plants evolved alternative photosynthesis types. One alternative, C4 photosynthesis, independently evolved at least 62 times in 19 different families of flowering plants. Plants with the C4 trait intensify their carbon fixation by using a biochemical pump to increase the concentration of CO2 at the site of Rubisco. As a result, C4 plants, such as maize, are known to have high growth rates. In a recent project, two researchers from the Bielefeld University and the IPK in Gatersleben applied constraint-based modeling (CBM) in order to find out which selective pressures lead to the evolution of the C4 pathway. 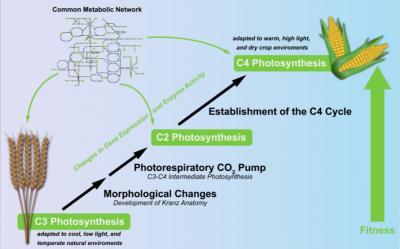 {module INSIDE STORY}
{module INSIDE STORY}
CBM enables researchers to apply different physical, enzymatic, and topological constraints when modeling metabolic networks. Therefore, different computational predictions can be made for a row of differing experimental scenarios, letting scientists glimpse the various possible routes evolution took or could have taken, depending on the settings of the constraints. After developing their C4-CBM-model the two scientists, Prof. Andrea Bräutigam, professor for computational biology at Bielefeld's Center for Biotechnology (CeBiTec) at Bielefeld University, and Dr. Mary-Ann Blätke, a member of the Network Analysis and Modelling IPK research group, focused on finding the constraints which led to the prediction of C4 photosynthesis as the optimal solution. Prof. Bräutigam: "Once the models are set up, observation of in silico evolution becomes possible. In our case, the simulations reproduced the evolutionary trajectory from C3 to C4 photosynthesis, which depended on the carbon dioxide level." Dr. Blätke complemented: "The model also predicts intermediacy as an optimal solution under particular conditions and explains why different variations of C4 photosynthesis may exist. It also put forth nitrogen and light as new eco-physiological parameters that play a role for the evolution of C4 photosynthesis."
The study showcases CBM as a powerful tool for querying and understanding the evolution of other complex traits in plants. Simultaneously, the successful analysis of the C4 evolution paves the way for the more detailed investigation of the C4 evolution and metabolism but also highlights new targets for future breeding and engineering efforts in C4-crop plants. Dr. Blätke: "A metabolic network correctly predicting the trajectory of C4 evolution, such as the one provided here, is a prerequisite to approach more detailed questions on C4 metabolism and its evolution. It can, therefore, be used as a working horse for follow-up studies and act as an integrative framework for multi-omic data and derived regulatory networks."


 How to resolve AdBlock issue?
How to resolve AdBlock issue?