Researchers from Utrecht University, one of the oldest universities in the Netherlands, have visualized the "molecular postman" that makes sure that many important proteins are delivered to the outside of the cell. This has profound implications for our understanding of protein secretion, which in the future can be used for the production of cheaper protein-based therapeutics such as insulin, and the development of new drugs against Corona-, Dengue-, and Zika-viruses as well as novel antibiotics. The researchers are publishing their results today in the academic journal Molecular Cell.
Many proteins, like antibodies, hemoglobin, and some hormones, are produced on the inside of cells and must subsequently be trafficked to the exterior so they can function properly. Like in a post office, cells attach ‘address labels’ called signal peptides to these protein molecules to send them where they are needed. However, in a ‘cellular post office’, some of these address labels need to be removed for the cargo to function properly, while some others must remain, with equally detrimental consequences in case of failure. Therefore, the distinction between these cases is crucial for the function of all sorts of life-sustaining molecules. 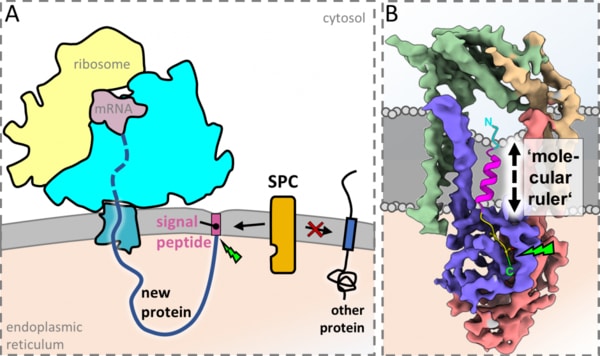
Molecular postman
Using a Nobel prize-winning, state-of-the-art imaging technique called cryo-electron microscopy (cryo-EM), the team of scientists lead by Prof. Friedrich Förster, in collaboration with Prof. Richard Scheltema and his group, has now visualized the human Signal Peptidase Complex (SPC), the ‘molecular postman’ that is at the center of this decision-making process and has to process more than 3,000 different cargo proteins. The research has revealed that the SPC uses a ‘molecular ruler’ made of phospholipids to measure the length of the signal peptides to decide which address labels to remove and which ones to keep.
Formidable challenge
Lead author Manuel Liaci explains: “Visualising the molecular architecture of the SPC was a formidable challenge that had been unmet for decades. The SPC is extremely small and it is integrated into the membrane of the cell, which makes it incredibly difficult to look at with the sub-nanometer resolution we needed.” Prof. Förster and his team employed a multidisciplinary approach combining their imaging technique with mass spectrometry and molecular dynamics simulations on a supercomputer, which enabled them to precisely define the molecular mechanism of signal peptide recognition and removal. The revelation of the SPC structure represents a major advancement in the field since it is the smallest membrane protein that has been visualized to near-atomic detail by cryo-EM so far.


 How to resolve AdBlock issue?
How to resolve AdBlock issue?