MOVIES
The Pieces of the Pathology Puzzle
Keeneland's GPU–CPU Combination Facilitates Large-scale Image Analysis in a Multi-faceted Approach to Understanding Disease
As disease progresses over space and time in the body, high-resolution imaging can capture the changes taking place down to the sub-cellular level; meanwhile, huge sets of hereditary (genomic) information hold clues about the dynamics of illness. Comparing certain characteristics in the images with genomic and clinical data may be key in predicting disease progression and in targeting new treatments. The current work of a research team from Emory University in Atlanta and Oak Ridge National Laboratory (ORNL) revolves around making those very connections.
The researchers are using high-resolution images of thin slices of brain tumors called gliomas in the comparisons to understand what’s involved in the onset and progression of the tumors. The knowledge acquired will allow for the prediction of disease behavior and the targeting of new treatments.
The overall emphasis of the project, however, is not specific to brain tumors but to understanding the relationships among disease morphology in general, genomic information and clinical outcomes in various ailments while also advancing computer science by optimizing the use of emerging hybrid supercomputer systems that have both central processing units (CPUs) and graphics processing units (GPUs).
Joel Saltz, professor and chair of Biomedical Informatics and director of the Center for Comprehensive Informatics (CCI) at Emory University, is leading the research team, which is composed of experts in pathology (the study of the nature and causes of diseases), microscopy imaging (the use of microscopes), image analysis, and high-performance computing (HPC). The Keeneland project is providing the HPC resources needed for large-scale image analysis.
Keeneland is a National Science Foundation-funded partnership between Georgia Tech, the National Institute for Computational Sciences (NICS), ORNL, the University of Tennessee, NVIDIA and Hewlett-Packard created to enable large-scale computational science on heterogeneous architectures in a coordinated manner. NICS provides the Keeneland project with HPC systems administration, file systems, high-performance wide-area-network connectivity, front-line user support and advanced user assistance.
Information from radiology, microscopy imaging, and genomic data holds clues to the mechanisms of disease, since the effects of disease generally manifest themselves as changes at the molecular, micro-anatomic and macro-anatomic levels. Micro-anatomicrefers to biological entities at the cell level, and macro-anatomic pertains to larger structures that can be captured in radiology images.
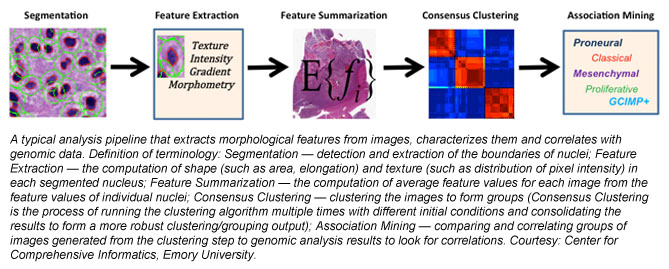
Research team member Tahsin Kurc of CCI explains that he and his colleagues are developing a combination of image analysis techniques and machine-learning-based (artificial intelligence) classification methods to glean morphological information at the cellular and sub-cellular scale from high-resolution images of tissue specimens. “Integration and correlation of this information with radiology, genomic and clinical information has shown tremendous potential to better understand the mechanisms of onset and progression of brain tumors,” Kurc says.
The set of medical images for analysis has been growing in recent years as high-resolution tissue scanners (a form of advanced digital camera), once prohibitively expensive, have become much more affordable, presenting the impetus to process the data. Says Kurc: “Now a 50,000- by 50,000-pixel image can be captured in a few minutes. The computational problem is that we have 5,000 ‘high-rez’ images, ranging from 20,000 pixels by 20,000 pixels to 100,000 pixels by 100,000 pixels each.”
Kurc explains that Keeneland allows for the analysis of a large number of the high-resolution images quickly, and that what used to take weeks can now be done in a few minutes. Keeneland features a 3:2 ratio of GPUs to CPUs and capitalizes on the ability of GPUs to process data up to 80 times faster than CPUs, he adds. The researchers are developing algorithms to avoid idle CPUs or GPUs during data processing, thus optimizing the use of Keeneland’s hybrid GPU–CPU system and contributing to the body of computer-science knowledge.
To take advantage of Keeneland, the research team has implemented GPU-enabled versions of the image-processing operations in the image-analysis pipelines and developed a software platform to execute the pipelines on multiple nodes of Keeneland and multiple CPUs and GPUs. In the context of this research,pipeline refers to a series of various operations to facilitate the analysis of an image.
“Keeneland has multi-core CPUs and multiple GPUs on each computation node [connection point],” Kurc says. “By carefully scheduling the operations to CPU cores and GPUs and distributing computation across multiple nodes, we are able to analyze one hundred and fifty 4,000 pixels by 4,000 pixels image tiles per second on 100 nodes of Keeneland.”
The pieces of the puzzle are coalescing to provide clues about brain tumors. Kurc explains the degree of progress in the research so far: “Our work using brain-tumor-tissue images from the Cancer Genome Atlas repository shows that the morphological characteristics of brain cancer tumors have good correlations to observed patient survival, indicating prognostic value of disease morphology. Our work has also demonstrated that grouping of tumors into prognostically significant clusters based on morphology is also correlated with variations in pathology and genetics and cancer-related pathways. These results show great potential in morphology analysis as a complementary platform for studying disease mechanisms and subtypes. Cancer tumors are heterogeneous, containing a mixture of cell types that may have different roles in sustaining the tumor and facilitating tumor growth. More research is needed to develop enhanced models of disease morphology to capture, represent and interpret this heterogeneity and to understand its relationship to patients' genomic profiles and clinical outcomes.”
