SCIENCE
Using systems biology to reveal the determinants of cell fate
Proliferation, differentiation or death? How is the fate of a cell determined? This system is a fundamental phenomenon in living organisms, but it has not yet been analyzed. Mariko Okada, team leader of the Laboratory for Cellular Systems Modeling at the RIKEN Research Center for Allergy and Immunology (RCAI), is working to analyze the fate-determining mechanisms in cells using systems biology, a new field of science in which experiments and theory are combined. She recently succeeded in revealing a molecular network that appears in cell differentiation processes and identified incoming information that triggers cell differentiation, thus establishing a model for mathematical prediction of input to output. The results can be used in various applications, such as supersupercomputer-aided searches for drug candidates and measurement of their effects.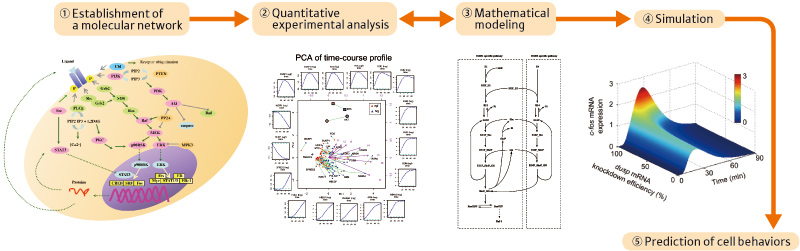
The determination of cell fate
“The cellular world resembles human society,” says Okada. “A cell contains various molecules such as proteins, including enzymes and growth-promoting substances as well as nucleic acids—DNA and RNA. Molecules have likes and dislikes; they combine or react with specific molecules. However, their preferences vary depending on the surrounding circumstances, such as the molecular shapes of their counterparts. I believe we all experience something similar.”
“I want to know how the fate of a cell is determined,” says Okada. Each cell that constitutes an individual living organism has DNA with exactly the same base sequence as every other cell; that is, the same set of DNA. However, cells differentiate, proliferate, or die depending on their sites, the point in time, and the surrounding circumstances. “What determines the fate of a cell is basically the preferences of its molecules.”
Many types of receptor are embedded in the cell membrane that covers a cell. When molecule A binds to a certain receptor, the information that “molecule A has bound to the receptor” is sent into the cell. Intercellular molecule B then reacts with molecule C, which in turn reacts with molecule D and so on, causing a chain reaction. The information is finally sent into the nucleus, leading to gene expression. The fate of a cell is thus determined by the amount and types of genes expressed in the cell.
“Many molecules involved in the information transmission process up to the determination of the fate of the cell have been discovered. However, when, where, and how these molecules work is not well understood. In addition, almost the same molecules involved can lead to different fates. I believe that not all the molecules involved in the transmission of information play major roles, but that there is a core group of molecules that play an important role in cell fate determination. I really want to provide a convincing explanation of the fate-determining system of the cell.”
Encountering systems biology
Before joining RIKEN, Okada conducted research on cell biology at the University of California, Davis in the US. “I originally majored in biochemistry and conducted research on enzymes both at graduate school and at the company I worked for. Enzymes have definite likes and dislikes and combine only with specific molecules, changing their chemical properties. Some enzymes, when combined with another molecule such as a co-enzyme or subunit, can accelerate chemical reactions. To understand cell phenomena, it is essential to know the behavior of these individual molecules. I was not completely satisfied with the research on cell biology and I was not provided with the necessary research tools,” says Okada, reflecting on days past.
After returning home in 2000, Okada joined the RIKEN Genomic Sciences Center and became a research scientist on the Computational Genomics Team in the Bioinformatics Group. “Before joining the team, the only experience I had was doing experiments on living organisms. However, I found that most of my fellow scientists were information science researchers. It was while I was thinking about what to do here in this team that I found a paper written by Boris Kholodenko.”
Now, Kholodenko is the deputy director of Systems Biology at University College Dublin in Ireland. He published a paper in 1999 on the ‘quantification of short-term signaling by the epidermal growth factor receptor’. “A cell begins to proliferate when epidermal growth factor binds to a receptor. Doctor Kholodenko described the intermolecular reactions during the information transmission process that occurs in a cell as a combination of differential equations and established a mathematical model. Aiming to establish a prediction system, he considered the information on a molecule binding to a receptor as an input and the information on gene expression as an output. He viewed the process from input to output as a prediction system. This paper proposed a novel idea at that time because it dealt not only with theory, but also with experiment.”
Researchers in life sciences have recently been paying increasing attention to systems biology. They are striving to find effective means for viewing individual life phenomena as a system to clarify their working principles. The general research process in systems biology proceeds as follows (Figure 1):
(1) Extraction of genes involved in target life phenomena or interactions between proteins from papers and experimental data to establish a molecular network.
(2) Comprehensive chronological and quantitative measurements of individual expression levels of genes, and concentrations and distributions of proteins by experiment.
(3) Mathematical analysis of huge amounts of data to establish a molecular network.
(4) supercomputer simulation under various conditions and comparison with experimental results to study the working principles and control system of life phenomena.
(5) Clarification of the relationship between input and output information to predict how a cell responds to the input and to identify input conditions that lead to a desirable output, so that the results can be applied to drug discovery.
The paper by Kholodenko is regarded as a precedent for systems biology. “On reading his paper, I thought, ‘This is it!’ Soon I started research activities modeled on his example. I felt that I could, but this was more than I could have dreamed of,” says Okada with a smile on her face.
Cell differentiation-inducing systems
Aiming to advance her own research into the fate-determining systems of the cell using systems biology, in which experiment and theory are combined, Okada started the Laboratory for Cellular Systems Modeling at the RCAI in 2008. In May 2010, one of her research results was published in the journal Cell. “This research was conducted in collaboration with Dr Kholodenko. In this paper, we used a breast cancer cell line called MCF-7 and successfully established a mathematical model that can simulate the differentiation-inducing network of the cell.”

Figure 2: Molecular network inducing differentiation in breast cancer cells.
(1) First, heregulin binds to a receptor on the cell membrane of the cell, which leads to the activation of ERKs through multiple molecules.
(2) An activated ERK moves into the nucleus of the cell, and in turn activates RSK.
(3) Both the ERK and RSK activate a transcription factor and express the fos gene.
(4) The fos RNA is carried to the cytoplasm to form FOS protein.
(5) The FOS protein is activated and then moves into the nucleus to express the gene and induce differentiation. The AND gate controls fos gene expression. In addition, the AND gate and the feed- forwarding exercise double control over the activation of FOS protein.
To be more precise, epidermal growth factor that promotes cell proliferation and heregulin, a molecule that promotes cell differentiation, were administered to breast cancer cells. Measurements of the concentration or distribution of proteins related to proliferation and differentiation, the speed of association with or dissociation from other proteins, and gene expression were taken from every few minutes to every few tens of minutes. The measured data were analyzed to create a mathematical model, which was then used for supercomputer simulation under various conditions, comparisons between the calculated and experimental results, and verification of the mathematical model. As a result, she successfully clarified a molecular network that worked only when the breast cancer cell differentiated, as well as the control system for the molecular network.
Figure 2 shows the differentiation-inducing molecular network that she clarified this time. When heregulin binds to a receptor on the cell membrane, the information is sent to an ERK molecule through many molecules. The ERK molecule then moves into the nucleus to activate RSK. Both ERKs and RSK activate a transcription factor (protein that binds to a specific DNA sequence and controls gene expression), which in turn promotes expression of the gene called fos. The fos RNA is then carried to the cytoplasm, where the FOS protein is produced. This FOS protein is also a transcription factor; when activated, it moves to the nucleus to express differentiation-inducing genes.
This is the ingenious way in which the differentiation-inducing system is controlled. The fos gene expresses itself only when information is received from both ERK and RSK at the same time through a transcription factor. Here, the control logic that is turned on and produces an output only when two information inputs are provided is known as an ‘AND’ gate. There are various sources of noise in a cell, which may cause improper operating signals when a switch is turned on by a single input. Introduction of an AND gate thus avoids the occurrence of malfunction and helps ensure a stable output.
“FOS protein activation is also controlled by an AND gate, because FOS protein is activated only when both the fos gene and ERK activity exist in the cytoplasm at the same time. We also found that the input from ERK in the cytoplasm was controlled by a ‘feed-forward’ loop.”
With most transmission of information, the information from a molecule is sent only to the molecule located next to that molecule. With feed-forward-based information transmission (Figure 2), on the other hand, information from the molecule is sent not only to the molecule located next to that molecule, but also to the molecule located several steps ahead. Feed-forward and the AND gate are general strategies used in control engineering, especially to prevent malfunction due to noise. They can also respond to the constantly changing cellular environment.
“Activation of ERKs that have moved to the nucleus from the cytoplasm has been known to last only several minutes, whereas activation of ERKs in the cytoplasm continues, although the roles of the ERKs have remained unclear. This time, however, we have clarified that ERKs play a role in feed-forward control.”
The production of FOS protein requires about 30 minutes after the initial entry of information from ERK into the nucleus. If the activation of ERK continues in the cytoplasm, double control by feed-forward and the AND gate results in activation of the FOS protein, leading to cell differentiation. However, the situation around the cell is changing from moment to moment. If activation of ERKs in the cytoplasm is stopped for some reason, the AND gate cannot be turned on because there is no incoming feed-forward information. As a result, the FOS protein is not activated and decomposes, resulting in the inhibition of cell differentiation.
“We have shown that the cell differentiation-inducing system consists of various control processes nested in several steps. It is the rigorous control of these control processes that converts analog information such as changes in the concentration of molecules into digital information to determine whether to differentiate or not. Viewing this molecular network model, I felt that it was great and beautiful.”
After publishing the paper, Okada received many praising emails from overseas researchers. There are several reasons for such a positive response. “The cell differentiation system we have clarified this time seems to be the common basic system, not only for the breast cancer cells we actually experimented on, but also for other types of cells. In other words, our model provides a useful means for computer simulation of differentiation in various cells types. The process can also be applied to selecting drug candidates and predicting their effects,” says Okada.
Overseas drug companies and venture companies are also interested in this paper. “The output from the AND gate changes if one of the inputs is inhibited. This fact means that the cell may proliferate rather than differentiate. If we can develop techniques to control cell differentiation and proliferation, to differentiate cells into the desired types of cells, and furthermore, if we can get them to proliferate as we want, the techniques will also contribute to regenerative medicine.”
Okada is offering the published molecular network model for any researcher to use freely. “If other researchers actually use the model and improve on the defects, the molecular network model will improve and will in turn be used by other researchers, thus producing numerous results. Knowledge-sharing is crucial to the development of science.”
The next target is immunity
“The term ‘systems biology’ sounds attractive, but it requires enormous and persistent efforts,” says Okada. “There are virtually no researchers who are talented in both biology and information science at the same level. For example, I have trouble with mathematics. When I listen to conversations between researchers in biology and researchers in information science, I have trouble even understanding what the problem is. However, I know that we need to be patient when talking to one another.” Okada insists that what is required in systems biology is patience. “We need to conduct discussions and experiments using a lot of patience. Fortunately, I am a patient person.”
Okada’s next research subject will be immunity. “Doctor Masaru Taniguchi, director of the RCAI, has asked me to use systems biology to explain immunological diseases. This subject is our long-term target. It is a challenging theme, but we believe that the approach we used for breast cancer cells can also be applied to immune cells. Since cultured cells are frequently used in experiments on systems biology, we often hear the criticism that whether you can apply results from experiments on cultured cells to actual biological systems to explain biological phenomena is questionable. I therefore want to use experimental data at the individual level to bridge the gap between the model and the real world. First of all, I want to produce some significant results within five years.
“I want to logically understand the fate-determining system of the cell, but humans tend to believe in their intuitive understanding. They want to follow their intuition while using highly complicated experiments and mathematical analysis. This fact contains a contradiction, but I am interested in it precisely because of the contradiction.”

