Constructing a mathematical basis for developing therapeutic strategies against cancer using a supercomputer
Understanding the principles of cancer evolution is important in designing a therapeutic strategy. A research group at The Institute of Medical Science, The University of Tokyo (IMSUT) announced a new simulation model that describes various modes of cancer evolution in a unified manner.
"We clarified the conditions under which each of the evolutionary modes is realized by performing simulation analysis using a supercomputer. Our findings have allowed us to explain the underlying principles of the evolutionary diversity of cancer." said lead scientist Atsushi Niida, Senior Assistant Professor, Laboratory of Molecular Medicine, Human Genome Center of IMSUT.
To build a unified evolutionary simulation model
Cancer can be regarded as a disease of evolution, which results from the natural selection of cells with high proliferative and malignant potential, following the accumulation of mutations in their genomes. Moreover, because cancer has a high evolutionary potential, it easily adapts to treatment-related changes in its environment and acquires therapeutic resistance. 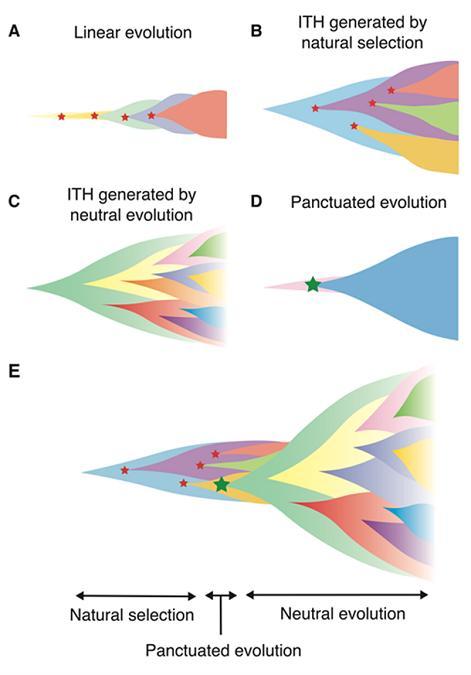 {module INSIDE STORY}
{module INSIDE STORY}
Previous genomic studies have shown that cancer evolution can be roughly divided into four modes of evolution. However, it has remained unclear what conditions give rise to each mode. Therefore, the research group has built a unified evolutionary simulation model that can recapitulate a variety of evolutionary modes.
By performing massively parallel simulation with various conditions on SHIROKANE, a supercomputer at IMSUT, the research group determined the conditions under which each evolution mode is generated. By providing a mathematical basis for understanding cancer evolution, this study is expected to contribute to the understanding of therapeutic resistance in cancer and the development of novel therapeutic strategies.
Various mathematical principles that explain the evolution of cancer
Classically, cancer evolution has been regarded as a process of evolution, in which normal cells evolve step-wise into a homogeneous cell population with high malignancy by serially acquiring driver mutations that favor cell growth and survival while undergoing natural selection.
However, in recent years, multi-region sequencing, which analyzes DNA samples obtained from multiple regions of a tumor, has demonstrated intratumor heterogeneity (ITH); that is, multiple clones with different mutations are generated during the course of cancer evolution and coexist in a single tumor. Also, depending on the types of cancers, driver mutations present only in a fraction of cells contribute to ITH; that is, natural selection appears to shape ITH.
On the other hand, it has been found that ITH can be shaped by the accumulation of neutral mutations that do not affect the growth and survival of cells; that is, ITH can be generated by neutral evolution. The evolutionary principle underlying ITH has not only varied among cancer types, but Niida and colleagues have also found that a temporal shift occurs during colorectal tumorigenesis (Saito et al.,2018); that is, it has been found that, in early lesions, ITH is shaped by natural selection while in advanced cancer, ITH is shaped by neutral evolution. Also, in contrast to the gradual evolution that results from the stepwise accumulation of single nucleotide mutations as assumed above, punctuated evolution has attracted attention. This is an evolutionary model in which large changes occur over a short time at the chromosomal and genomic levels, such as copy number changes and structural abnormalities, explosively expanding from a few initial cells.
Thus far, four evolutionary modes have been proposed, but there are many unclear points concerning under what conditions these modes occur. Simulation using an agent-based model is considered to be a useful tool for understanding the principle of cancer evolution. The agent-based model assumes the components of a system called an agent and specifies rules for the autonomous behavior of the agent itself and the interactions between agents and the environment.
Viewing each cell as an agent, ITH can be easily represented by the difference in the internal state of each agent. Niida and colleagues have recently developed MASSIVE, a new methodology of parameter sensitivity analysis, which examines the dynamics of agent-based simulations (Niida et al., 2019). MASSIVE takes an approach completely different from conventional parameter sensitivity analysis methods; it makes it possible to intuitively search a large parameter space by combining massively parallel computing with interactive data visualization.
Based on the above, in order to clarify the conditions under which the above-mentioned four different evolution modes are realized, we have constructed a unified evolution simulation model that can recapitulate various evolution modes using an agent-based model. Parameter sensitivity analysis utilizing MASSIVE was performed on SHIROKANE, a supercomputer of IMSUT. They found that linear evolution occurs when strong driver mutations are assumed, while ITH is generated by natural selection when the driver mutations are weak. Moreover, the simulation revealed that the generation of ITH by neutral evolution requires a high neutral mutation rate, and the presence of cancer stem cells also contributes to neutral evolution by promoting the accumulation of neutral mutations.
Punctuated evolution could also be reproduced by assuming an explosive driver gene that enables cells to overcome the resource limitation required for cell proliferation. Furthermore, it was shown by simulation that punctuated evolution triggers the above-described temporal shift of the principle underlying ITH from natural selection to neutral evolution in colorectal tumorigenesis (Fig. 1E). This result also helps us understand that each mode works not as discrete, exclusive modes but rather blend continuously as a series of phases of cancer evolution.
The results of all the simulation analyses in this research can be searched interactively at https://www.hgc.jp/~aniida/canevosim/index.html.
Contributing to a deeper understanding of therapeutic resistance in cancer
"In this study, we clarified the principle underlying the diversity of cancer evolution by simulation analysis using a supercomputer. Since cancer has a high evolutionary potential, it adapts to the environment changed by therapy and easily acquires therapeutic resistance, so understanding the evolutionary principle of cancer is an important problem for designing therapeutic strategies." Niida emphasized
By providing a mathematical basis for understanding cancer evolution, this study is expected to contribute to the understanding of therapeutic resistance in cancer and the development of novel therapeutic strategies.


 How to resolve AdBlock issue?
How to resolve AdBlock issue?