SYSTEMS
Meet the Gribbles
Understanding a Novel Enzyme from a Family of Marine Crustaceans May Bolster Biofuels Development
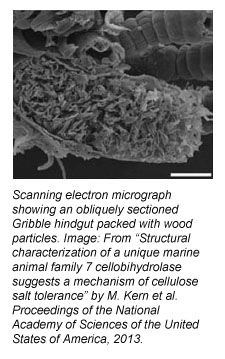
Tiny, wood-boring marine crustaceans with a funny name and a penchant for collectively attacking piers and dining on driftwood, ships, boats and docks have made a big splash in the science news media lately. These creatures, called Gribbles (scientific name: Limnoria quadripunctata), have as their recent claim to fame, a novel biomass-degrading enzyme in their guts that is of keen interest to the biofuels research and engineering communities.
Biomass from waste plant materials can be used to make biofuels when its main structural component, cellulose, is depolymerized — that is, broken down — and its component sugar, glucose, is fermented into ethanol or converted into other transportation fuels such as gasoline, diesel or jet fuel. However, low-cost, efficient enzymes tough enough to withstand industrial conditions are needed to enhance the economic viability of processes to produce biofuels, alternatives to fossil fuels.
Novel enzymes found in the wood-eating Gribbles, which hail from intertidal zones of the ocean, may provide clues on how to meet those needs and bolster the biomass-conversion process by enabling the creation of tougher enzymes, as some of the enzymes found in the Gribble are of the same family as ones almost exclusively found in fungi, but with several novel properties.
“Typically, in the development of biomass degrading enzymes, fungi are the primary organisms of interest from which to harvest enzymes, as fungi are responsible for the majority of biomass degradation in nature,” says Gregg Beckham of the U.S. Department of Energy’s National Renewable Energy Laboratory (NREL). “They turn over huge amounts of plant material, across a huge range of natural environments. So fungi are traditionally a good place to start when looking for enzymes for industrial use.”
Many studies have been focused on fungal enzymes as well as understanding enzymes in creatures that obviously eat biomass for a living, such as cows or termites. But Beckham explains that Gribbles represent something entirely different from those usual subjects in that they don’t rely on gut bacteria to make enzymes to aid their digestion. Their guts are sterile (free of bacteria) and the organism produces enzymes itself in an organ called the heptopancreas, which runs the length of their body.
Beckham is a member of a team of researchers composed of members from the United Kingdom, NREL and the University of Kentucky who have recently published a paper describing the crystal structure of a unique endogenous (made by the organism itself) cellulase (cellulose-degrading enzyme) that Gribbles produce. The paper appears online in PNAS, the Proceedings of the National Academy of Sciences of the United States of America, and is entitled, “Structural characterization of a unique marine animal family 7 cellobiohydrolase suggests a mechanism of cellulose salt tolerance.” Britain’s Biotechnology and Biological Science Research Council (known as BBSRC) and the U.S. Department of Energy’s BioEnergy Technologies Office are funding the U.K. and U.S. work in the project, respectively.
Background: Research Leading to the Current Findings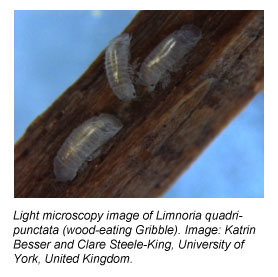
Beckham explains that his colleagues from the U.K. have spent several years conducting the research that ultimately led to the discovery and characterization of the novel Gribble enzyme. Professor Simon Cragg of the University of Portsmouth has long studied the zoology of Gribbles and other marine wood-boring crustaceans in intertidal zones.
While professors Cragg and John McGeehan at Portsmouth provide the zoology and structural biology expertise, respectively, researchers in York, led by professors Neil Bruce and Simon McQueen-Mason, have examined the transcriptome of the Gribble to determine which enzymes are secreted by Gribbles when fed wood, as well as biochemical characterization of the resulting individual, isolated enzymes.
In 2010, the researchers from the U.K. published the paper, “Molecular insight into lignocellulose digestion by a marine isopod in the absence of gut microbes,” also in PNAS, in which they described the transcriptome of L. quadripunctata. A transcriptome is the set of all the ribonucleic acid (RNA) produced in one or a population of cells. The paper reported an exciting finding: L. quadripunctata produces an enzyme from a well-known, industrially relevant family of cellulases called Family 7, typically found only in fungi.
“Family-7 enzymes are often the cornerstone of industrial enzyme cocktails for biomass conversion, and so that’s why the discovery was so important and interesting,” Beckham explains. In addition, the U.K. researchers discovered that the Gribble actually has three Family-7 enzymes, and one of those, called Cel7B, is the focus of their current investigation.
The Promising Novel Gribble Enzyme Cel7B
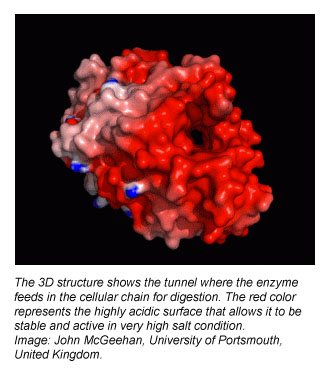
The U.K. researchers isolated the Gribble enzyme Cel7B and solved its structure with X-ray diffraction led by McGeehan. The York team characterized Cel7B to determine its activity at various conditions of interest. NREL was enlisted to perform molecular dynamics (MD) simulations, enabled by high-performance-computing (HPC) resources. The details of their characterization of Cel7B are contained in the current PNAS paper.
Beckham says: “One of the things that we immediately noticed when we were constructing the enzyme to run simulations was that the charge on the enzyme’s surface was immense. High negative surface charge is typically correlated with salt tolerance, a characteristic that may suggest the enzyme will be hardy in high solids, industrial environments. When we examined sequences and structures of known fungal enzymes from this family — and there are many hundreds of them known — we found that this feature is rather unique.”
Halophilic (“salt-loving”) organisms possess myriad proteins with high negative surface charges, and so in that context, the nature of the enzymes found in the Gribble’s gut makes perfect sense. Beckham says the researchers’ bench tests of Cel7B (detailed in the current PNAS paper) showed it to remain active at more than six times the salt concentration of the sea and even become slightly more effective in its ability to degrade biomass. Beckham explains further: “That led us to postulate in this paper that we can learn some things from this enzyme about engineering industrial enzymes for biomass conversion.”
A key driver in the investigation is finding an enzyme that will function effectively under the condition of “high solids loading” — that is, a high solids-to-water ratio. Beckham explains: “The less water you have in the process, the smaller your reactor can be. This concentrates the product (sugar), and saves on the cost of the reactor, other equipment and water.” The Cel7B enzyme, he adds, may provide clues as to how to design particular features of enzymes for enhanced stability for industrial application.
Structural Biology and MD Simulations
Characterizing the Gribble enzyme is crucial to understanding it, and the knowledge acquired could help in the design of a better enzyme for biomass degradation. Structural biology and MD simulation, in particular, complement each other quite nicely in the task of molecular-level characterization. “They work really beautifully together because structural biology gives you a static picture, and MD simulations can give you a dynamic picture,” Beckham says.
Beckham credits the MD simulations run using the HPC resources from the National Science Foundation’s eXtreme Science and Engineering and Discovery Environment (XSEDE) with enabling him and his research colleagues to make valuable inferences about enzymes and explain a number of experimental results.
“Kraken for us is a resource that’s incredibly useful for running long-production MD simulations, just like the Athena supercomputer before it,” Beckham says. “From this initial study, Kraken has already been essential for us to run additional simulations with this new enzyme structure. For example, the Cel7B enzyme has a very long tunnel that holds onto a cellulose chain extracted from a plant cell wall. The binding strength of the enzyme to the cellulose chain is likely a key metric for determining the enzyme’s effectiveness.”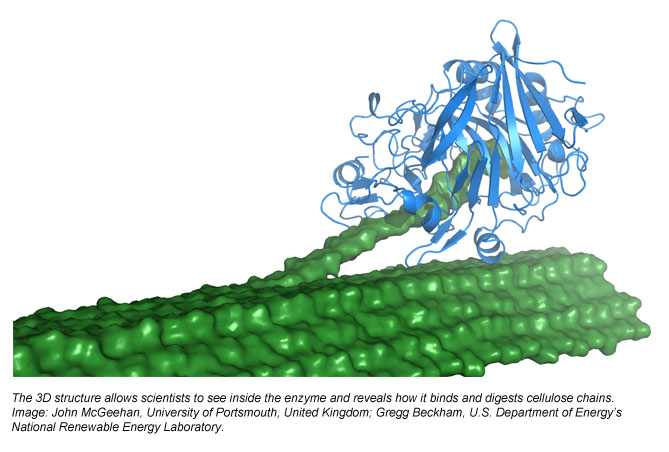
Going forward, the researchers are using Kraken to take the next step on this novel Family-7 enzyme from the Gribbles and compare it with conventional Family-7 enzymes. Beckham adds: "We will be able to use what we learn to make better predictions about enzyme activity and whether the enzyme can be used directly in biomass conversion or can be modified to be more like fungal enzymes while retaining useful characteristics, such as the ability for some salt and high-solids tolerance.”
Related Links
- "Enzyme from wood-eating gribble could help turn waste into biofuel," from the Biotechnology and Biological Science Research Council (BBSRC, United Kingdom)
- “Structural characterization of a unique marine animal family 7 cellobiohydrolase suggests a mechanism of cellulose salt tolerance,” paper in Proceedings of the National Academy of Sciences of the United States of America, 2013
- “Novel Enzyme from Tiny Gribble Could Prove a Boon for Biofuels Research,” news release from the National Renewable Energy Laboratory, June 11, 2013
- “Gribble enzyme might be holy grail for biofuel,” from UoP News (University of Portsmouth, United Kingdom), June 4, 2013
- “How the wood-eating gribble could help turn waste into biofuel,” from The University of York News and Events, June 4, 2013
